Back to Tutorials and Data Recipes
Authors: Sean Foley (NASA, MSU), Meng Gao (NASA, SSAI), Ian Carroll (NASA, UMBC)
The following notebooks are prerequisites for this tutorial.
An Earthdata Login account is required to access data from the NASA Earthdata system, including NASA ocean color data.
PACE has two Multi-Angle Polarimeters (MAPs): SPEXone and HARP2. These sensors offer unique data, which is useful for its own scientific purposes and also complements the data from OCI. Working with data from the MAPs requires you to understand both multi-angle data and some basic concepts about polarization. This notebook will walk you through some basic understanding and visualizations of multi-angle polarimetry, so that you feel comfortable incorporating this data into your future projects.
At the end of this notebook you will know:
Begin by importing all of the packages used in this notebook. If your kernel uses an environment defined following the guidance on the tutorials page, then the imports will be successful.
from pathlib import Path
from tempfile import TemporaryDirectory
from scipy.ndimage import gaussian_filter1d
from matplotlib import animation
import cartopy.crs as ccrs
import earthaccess
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
Download some HARP2 Level-1C data using the short_name
value “PACE_HARP2_L1C_SCI” in earthaccess.search_data.
Level-1C corresponds to geolocated imagery. This means the imagery
coming from the satellite has been calibrated and assigned to locations
on the Earth’s surface. Note that this might take a while, depending on
the speed of your internet connection, and the progress bar will seem
frozen because we’re only downloading one file.
auth = earthaccess.login(persist=True)
tspan = ("2024-05-20", "2024-05-20")
results = earthaccess.search_data(
short_name="PACE_HARP2_L1C_SCI",
temporal=tspan,
count=1,
)
Granules found: 1
paths = earthaccess.open(results)
Opening 1 granules, approx size: 0.62 GB using endpoint: https://obdaac-tea.earthdatacloud.nasa.gov/s3credentials
QUEUEING TASKS | : 0%| | 0/1 [00:00<?, ?it/s]
PROCESSING TASKS | : 0%| | 0/1 [00:00<?, ?it/s]
COLLECTING RESULTS | : 0%| | 0/1 [00:00<?, ?it/s]
prod = xr.open_dataset(paths[0])
view = xr.open_dataset(paths[0], group="sensor_views_bands").squeeze()
geo = xr.open_dataset(paths[0], group="geolocation_data").set_coords(["longitude", "latitude"])
obs = xr.open_dataset(paths[0], group="observation_data").squeeze()
The prod dataset, as usual for OB.DAAC products,
contains attributes but no variables. Merge it with the
“observation_data” and “geolocation_data”, setting latitude and
longitude as auxiliary (e.e. non-index) coordinates, to get started.
dataset = xr.merge((prod, obs, geo))
dataset
<xarray.Dataset> Size: 1GB
Dimensions: (bins_along_track: 396, bins_across_track: 519,
number_of_views: 90)
Coordinates:
longitude (bins_along_track, bins_across_track) float32 822kB ...
latitude (bins_along_track, bins_across_track) float32 822kB ...
Dimensions without coordinates: bins_along_track, bins_across_track,
number_of_views
Data variables: (12/20)
number_of_observations (bins_along_track, bins_across_track, number_of_views) float32 74MB ...
qc (bins_along_track, bins_across_track, number_of_views) float32 74MB ...
i (bins_along_track, bins_across_track, number_of_views) float32 74MB ...
q (bins_along_track, bins_across_track, number_of_views) float32 74MB ...
u (bins_along_track, bins_across_track, number_of_views) float32 74MB ...
dolp (bins_along_track, bins_across_track, number_of_views) float32 74MB ...
... ...
sensor_zenith_angle (bins_along_track, bins_across_track, number_of_views) float32 74MB ...
sensor_azimuth_angle (bins_along_track, bins_across_track, number_of_views) float32 74MB ...
solar_zenith_angle (bins_along_track, bins_across_track, number_of_views) float32 74MB ...
solar_azimuth_angle (bins_along_track, bins_across_track, number_of_views) float32 74MB ...
scattering_angle (bins_along_track, bins_across_track, number_of_views) float32 74MB ...
rotation_angle (bins_along_track, bins_across_track, number_of_views) float32 74MB ...
Attributes: (12/68)
title: PACE HARP2 Level-1C data
instrument: HARP2
product_name: PACE_HARP2.20240519T235950.L1C.5km.nc
processing_level: L1C
processing_version: V00
conventions: CF-1.10
... ...
geolocate_xtrack_amplification: 1.0
geolocate_atrack_amplification: 1.0
att_time_offset: 0.0
gps_time_offset: 0.0
harp_time_offset: 0.0
processing_status: CompletedHARP2 is a multi-spectral sensor, like OCI, with 4 spectral bands. These roughly correspond to green, red, near infrared (NIR), and blue (in that order). HARP2 is also multi-angle. These angles are with respect to the satellite track. Essentially, HARP2 is always looking ahead, looking behind, and everywhere in between. The number of angles varies per sensor. The red band has 60 angles, while the green, blue, and NIR bands each have 10.
In the HARP2 data, the angles and the spectral bands are combined into one axis. I’ll refer to this combined axis as HARP2’s “channels.” Below, we’ll make a quick plot both the viewing angles and the wavelengths of HARP2’s channels. In both plots, the x-axis is simply the channel index.
Pull out the view angles and wavelengths.
angles = view["sensor_view_angle"]
wavelengths = view["intensity_wavelength"]
Create a figure with 2 rows and 1 column and a reasonable size for many screens.
fig, (ax_angle, ax_wavelength) = plt.subplots(2, 1, figsize=(14, 7))
ax_angle.set_ylabel("View Angle (degrees)")
ax_angle.set_xlabel("Index")
ax_wavelength.set_ylabel("Wavelength (nm)")
ax_wavelength.set_xlabel("Index")
plot_data = [
(0, 10, "green", "^", "green"),
(10, 70, "red", "*", "red"),
(70, 80, "black", "s", "NIR"),
(80, 90, "blue", "o", "blue"),
]
for start_idx, end_idx, color, marker, label in plot_data:
ax_angle.plot(
np.arange(start_idx, end_idx),
angles[start_idx:end_idx],
color=color,
marker=marker,
label=label,
)
ax_wavelength.plot(
np.arange(start_idx, end_idx),
wavelengths[start_idx:end_idx],
color=color,
marker=marker,
label=label,
)
ax_angle.legend()
ax_wavelength.legend()
plt.show()
Both HARP2 and SPEXone conduct polarized measurements. Polarization describes the geometric orientation of the oscillation of light waves. Randomly polarized light (like light coming directly from the sun) has an approximately equal amount of waves in every orientation. When light reflects off certain surfaces or is scattered by small particles, it can become non-randomly polarized.
Polarimetric data is typically represented using Stokes vectors. These have four components: I, Q, U, and V. Both HARP2 and SPEXone are only sensitive to linear polarization, and do not detect circular polarization. Since the V component corresponds to circular polarization, the data only includes the I, Q, and U elements of the Stokes vector.
The I, Q, and U components of the Stokes vector are separate
variables in the obs dataset.
stokes = dataset[["i", "q", "u"]]
Let’s make a plot of the I, Q, and U components of our Stokes vector, using the RGB channels, which will help our eyes make sense of the data. We’ll use the view that is closest to pointing straight down, which is called the “nadir” view. It is important to understand that, because HARP2 is a pushbroom sensor with a wide swath, the sensor zenith angle at the edges of the swath will still be high. It’s only a true nadir view close to the center of the swath. Still, the average sensor zenith angle will be lowest in this view.)
The first 10 channels are green, the next 60 channels are red, and the final 10 channels are blue (we’re skipping NIR). In each of those groups of channels, we get the index of the minimum absolute value of the camera angle, corresponding to our nadir view.
green_nadir_idx = np.argmin(np.abs(angles[:10].values))
red_nadir_idx = 10 + np.argmin(np.abs(angles[10:70].values))
blue_nadir_idx = 80 + np.argmin(np.abs(angles[80:].values))
Then, get the data at the nadir indices.
rgb_stokes = stokes.isel(
{
"number_of_views": [red_nadir_idx, green_nadir_idx, blue_nadir_idx],
}
)
A few adjustments make the image easier to visualize. First, normalize the data between 0 and 1. Second, bring out some of the darker colors.
rgb_stokes = (rgb_stokes - rgb_stokes.min()) / (rgb_stokes.max() - rgb_stokes.min())
rgb_stokes = rgb_stokes ** (3 / 4)
Since the nadir view is not processed at swath edges, a better image
will result from finding a valid window within the dataset. Using just
the array for the I component, we crop the rgb_stokes
dataset using the where attribute and some boolean logic
applied across different dimensions of the array.
window = rgb_stokes["i"].notnull().all("number_of_views")
crop_rgb_stokes = rgb_stokes.where(
window.any("bins_along_track") & window.any("bins_across_track"),
drop=True,
)
The granule crosses the 180 degree longitude, so we set up the figure
and subplots to use a Plate Carree projection shifted to center on a
-170 longitude. The data has coordinates from the default (i.e. centered
at 0 longitude) Plate Carree projection, so we give that CRS as a
transform.
crs_proj = ccrs.PlateCarree(-170)
crs_data = ccrs.PlateCarree()
The figure will hav 1 row and 3 columns, for each of the I, Q, and U arrays, spanning a width suitable for many screens.
fig, ax = plt.subplots(1, 3, figsize=(16, 5), subplot_kw={"projection": crs_proj})
fig.suptitle(f'{prod.attrs["product_name"]} RGB')
for i, (key, value) in enumerate(crop_rgb_stokes.items()):
ax[i].pcolormesh(value["longitude"], value["latitude"], value, transform=crs_data)
ax[i].gridlines(draw_labels={"bottom": "x", "left": "y"}, linestyle="--")
ax[i].coastlines(color="grey")
ax[i].set_title(key.upper())
It’s pretty plain to see that the I plot makes sense to the eye: we can see clouds over the Pacific Ocean (this scene is south of the Cook Islands and east of Australia). This is because the I component of the Stokes vector corresponds to the total intensity. In other words, this is roughly what your eyes would see. However, the Q and U plots don’t quite make as much sense to the eye. We can see that there is some sort of transition in the middle, which is the satellite track. This transition occurs in both plots, but is stronger in Q. This gives us a hint: the type of linear polarization we see in the scene depends on the angle with which we view the scene.
This Wikipedia plot is very helpful for understanding what exactly the Q and U components of the Stokes vector mean. Q describes how much the light is oriented in -90°/90° vs. 0°/180°, while U describes how much light is oriented in -135°/45°; vs. -45°/135°.
Next, let’s take a look at the degree of linear polarization (DoLP).
rgb_dolp = dataset["dolp"].isel(
{
"number_of_views": [red_nadir_idx, green_nadir_idx, blue_nadir_idx],
}
)
crop_rgb_dolp = rgb_dolp.where(
window.any("bins_along_track") & window.any("bins_across_track"),
drop=True,
)
crop_rgb = xr.merge((crop_rgb_dolp, crop_rgb_stokes))
Create a figure with 1 row and 2 columns, having a good width for many screens, that will use the projection defined above. For the two columns, we iterate over just the I and DoLP arrays.
fig, ax = plt.subplots(1, 2, figsize=(16, 8), subplot_kw={"projection": crs_proj})
fig.suptitle(f'{prod.attrs["product_name"]} RGB')
for i, (key, value) in enumerate(crop_rgb[["i", "dolp"]].items()):
ax[i].pcolormesh(value["longitude"], value["latitude"], value, transform=crs_data)
ax[i].gridlines(draw_labels={"bottom": "x", "left": "y"}, linestyle="--")
ax[i].coastlines(color="grey")
ax[i].set_title(key.upper())
For a different perspective on DoLP, line plots of the channels averaged over the two spatial dimensions show the clear minimum associated with the nadir view angle.
dolp_mean = dataset["dolp"].mean(["bins_along_track", "bins_across_track"])
dolp_mean = (dolp_mean - dolp_mean.min()) / (dolp_mean.max() - dolp_mean.min())
fig, ax = plt.subplots(figsize=(16, 6))
wv_uq = np.unique(wavelengths.values)
plot_data = [("b", "o"), ("g", "^"), ("r", "*"), ("k", "s")]
for wv_idx in range(4):
wv = wv_uq[wv_idx]
wv_mask = wavelengths.values == wv
c, m = plot_data[wv_idx]
ax.plot(
angles.values[wv_mask],
dolp_mean[wv_mask],
color=c,
marker=m,
markersize=7,
label=str(wv),
)
ax.legend()
ax.set_xlabel("Nominal View Angle (°)")
ax.set_ylabel("DoLP")
ax.set_title("Mean DoLP by View Angle")
plt.show()
We can convert radiance into reflectance. For a more in-depth explanation, see here. This conversion compensates for the differences in appearance due to the viewing angle and sun angle.
The radiances collected by HARP2 often need to be converted, using additional properties, to reflectances. Write the conversion as a function, because you may need to repeat it.
def rad_to_refl(rad, f0, sza, r):
"""Convert radiance to reflectance.
Args:
rad: Radiance.
f0: Solar irradiance.
sza: Solar zenith angle.
r: Sun-Earth distance (in AU).
Returns: Reflectance.
"""
return (r**2) * np.pi * rad / np.cos(sza * np.pi / 180) / f0
The difference in appearance (after matplotlib automatically normalizes the data) is negligible, but the difference in the physical meaning of the array values is quite important.
refl = rad_to_refl(
rad=dataset["i"],
f0=view["intensity_f0"],
sza=dataset["solar_zenith_angle"],
r=float(dataset.attrs["sun_earth_distance"]),
)
fig, ax = plt.subplots(1, 2, figsize=(16, 8))
ax[0].imshow(dataset["i"].sel({"number_of_views": red_nadir_idx}), cmap="gray")
ax[0].set_title("Radiance")
ax[1].imshow(refl.sel({"number_of_views": red_nadir_idx}), cmap="gray")
ax[1].set_title("Reflectance")
plt.show()
Create a line plot of the mean reflectance for each view angle and spectral channel. The flatness of this plot serves as a sanity check that nothing has gone horribly wrong with our data processing.
fig, ax = plt.subplots(figsize=(16, 6))
wv_uq = np.unique(wavelengths.values)
plot_data = [("b", "o"), ("g", "^"), ("r", "*"), ("black", "s")]
refl_mean = refl.mean(["bins_along_track", "bins_across_track"])
for wv_idx in range(4):
wv = wv_uq[wv_idx]
wv_mask = wavelengths.values == wv
c, m = plot_data[wv_idx]
ax.plot(
angles.values[wv_mask],
refl_mean[wv_mask],
color=c,
marker=m,
markersize=7,
label=str(wv),
)
ax.legend()
ax.set_xlabel("Nominal View Angle (°)")
ax.set_ylabel("Reflectance")
ax.set_title("Mean Reflectance by View Angle")
plt.show()
WARNING: there is some flickering in the animation displayed in this section.
All that is great for looking at a single angle at a time, but it doesn’t capture the multi-angle nature of the instrument. Multi-angle data innately captures information about 3D structure. To get a sense of that, we’ll make an animation of the scene with the 60 viewing angles available for the red band.
We are going to generate this animation without using the latitude
and longitude coordinates. If you use XArray’s plot as
above with coordinates, you could use a projection. However, that can be
a little slow for all animation “frames” available with HARP2. This
means there will be some stripes of what seems like missing data at
certain angles. These stripes actually result from the gridding of the
multi-angle data, and are not a bug.
Get the reflectances of just the red channel, and normalize the reflectance to lie between 0 and 1.
refl_red = refl[..., 10:70]
refl_pretty = (refl_red - refl_red.min()) / (refl_red.max() - refl_red.min())
A very mild Gaussian filter over the angular axis will improve the animation’s smoothness.
refl_pretty.data = gaussian_filter1d(refl_pretty, sigma=0.5, truncate=2, axis=2)
Raising the image to the power 2/3 will brighten it a little bit.
refl_pretty = refl_pretty ** (2 / 3)
Append all but the first and last frame in reverse order, to get a ‘bounce’ effect.
frames = np.arange(refl_pretty.sizes["number_of_views"])
frames = np.concatenate((frames, frames[-1::-1]))
frames
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50,
51, 52, 53, 54, 55, 56, 57, 58, 59, 59, 58, 57, 56, 55, 54, 53, 52,
51, 50, 49, 48, 47, 46, 45, 44, 43, 42, 41, 40, 39, 38, 37, 36, 35,
34, 33, 32, 31, 30, 29, 28, 27, 26, 25, 24, 23, 22, 21, 20, 19, 18,
17, 16, 15, 14, 13, 12, 11, 10, 9, 8, 7, 6, 5, 4, 3, 2, 1,
0])
In order to display an animation in a Jupyter notebook, the “backend” for matplotlib has to be explicitly set to “widget”.
Now we can use matplotlib.animation to create an initial
plot, define a function to update that plot for each new frame, and show
the resulting animation. When we create the inital plot, we get back the
object called im below. This object is an instance of
matplotlib.artist.Artist and is responsible for rendering
data on the axes. Our update function uses that artist’s
set_data method to leave everything in the plot the same
other than the data used to make the image.
fig, ax = plt.subplots()
im = ax.imshow(refl_pretty[{"number_of_views": 0}], cmap="gray")
def update(i):
im.set_data(refl_pretty[{"number_of_views": i}])
return im
an = animation.FuncAnimation(fig=fig, func=update, frames=frames, interval=30)
filename = f'harp2_red_anim_{dataset.attrs["product_name"].split(".")[1]}.gif'
an.save(filename, writer="pillow")
plt.close()
This scene is a great example of multi-layer clouds. You can use the parallax effect to distinguish between these layers.
The sunglint is an obvious feature, but you can also make out the opposition effect on some of the clouds in the scene. These details would be far harder to identify without multiple angles!
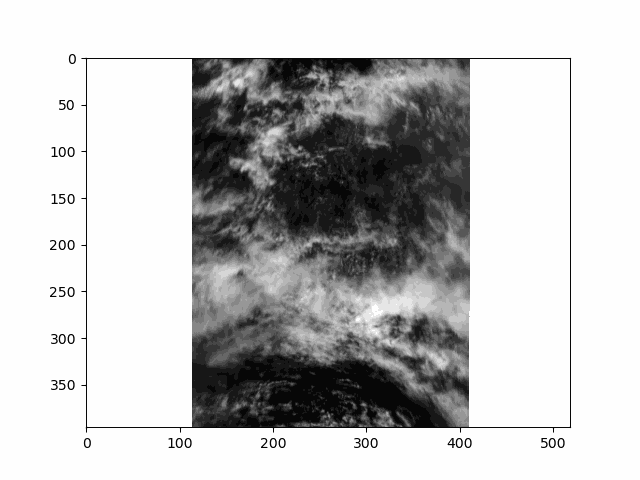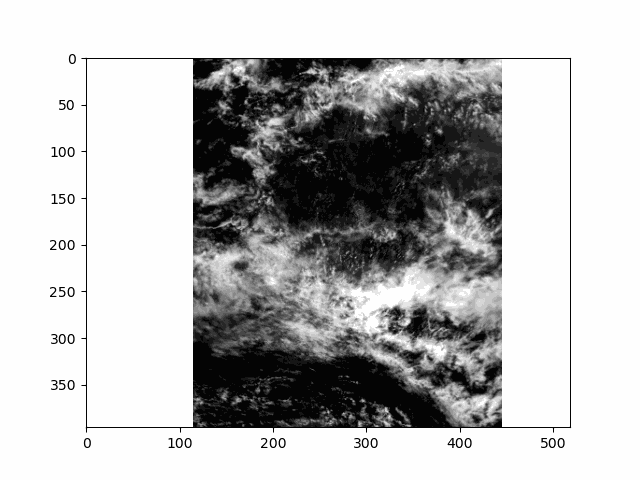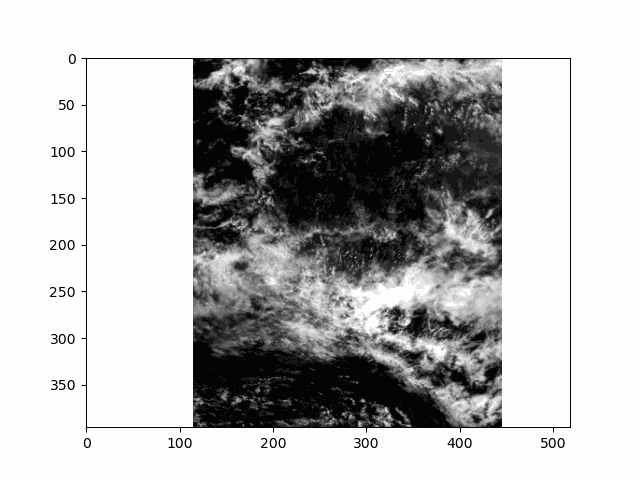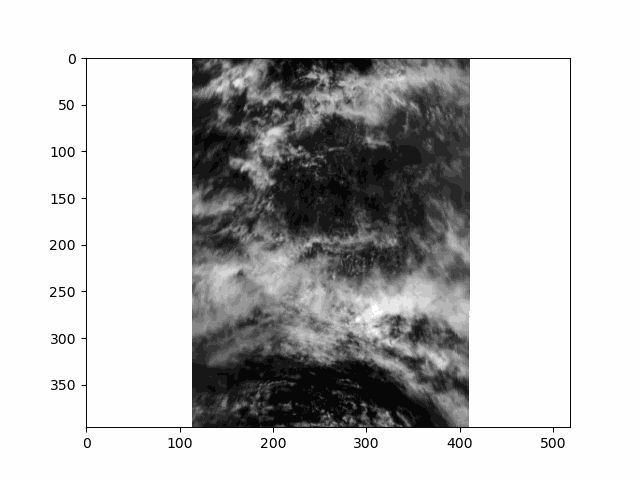
Notice the cell ends with plt.close() rather than the
usual plt.show(). By default, matplotlib will
not display an animation. To view the animation, we saved it as a file
and displayed the result in the next cell. Alternatively, you could
change the default by executing %matplotlib widget. The
widget setting, which works in Jupyter Lab but not on a
static website, you can use plt.show() as well as
an.pause() and an.resume().
You have completed the notebook giving a first look at HARP2 data. More notebooks are comming soon!